The following example will show how to compute CSA on T1w scans which were acquired for the brain, but where the upper cervical spinal cord is visible:
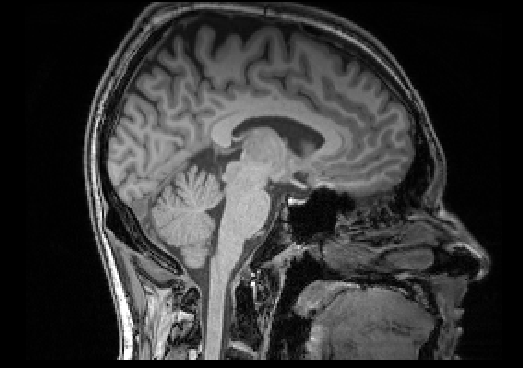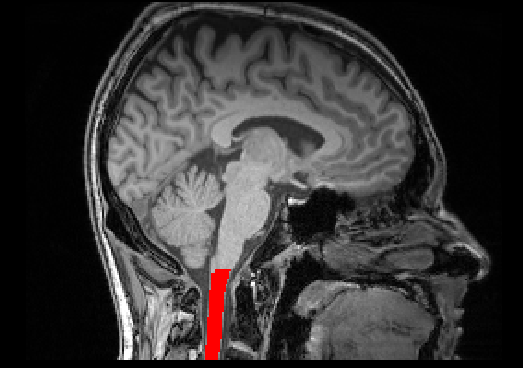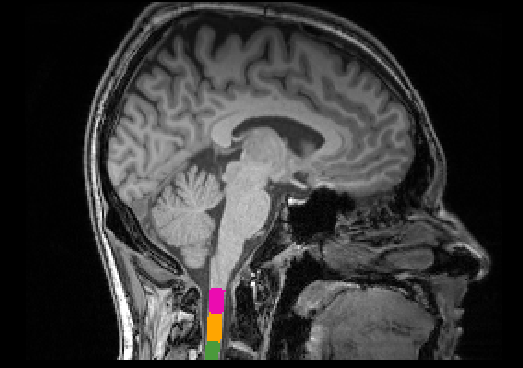
# This code has been tested with SCT v4.2.1
# Download example data
sct_download_data -d sct_example_data
# Crop to simulate a brain-only scan with the cord visible down to C2-C3 disc
sct_crop_image -i t1.nii.gz -zmin 125
# Segment the spinal cord, specifying that a brain is present in the image
sct_deepseg_sc -i t1_crop.nii.gz -c t1 -brain 1 -centerline cnn
# Label vertebrae
sct_label_vertebrae -i t1_crop.nii.gz -s t1_crop_seg.nii.gz -c t1
# Compute average CSA between C1-C2 level
sct_process_segmentation -i t1_crop_seg.nii.gz -vertfile t1_crop_seg_labeled.nii.gz -vert 1:2
Here is the output CSA (including other morphometric measures): csa.csv (777 Bytes)
Hi @jcohenadad - we are successfully running this on several (many!) brain datasets! However, some datasets seem to fail semi-randomly (based on visual QA - we cannot see differences in image field of view, quality, anatomy). Are there any tips/tricks/parameters that you may have learned in the last few years since this post? Admittedly, the datasets where we have “random” failures are lower overall quality, but it succeeds on other subjects within the same cohort! The failure occurs in the sct_deepseg_sc with:
EmptyArrayError: ‘Spinal cord not detected. Please make sure that there is sufficient contrast between the spinal cord and CSF to ensure good results.’
We do reorient to RPI, and resample to 1mm isotropic prior to sct_deepseg_sc. Example screenshots below so you can see data quality:
We suspect these are borderline quality/contrast for sct_deepseg_sc.
Thank you,
Kurt
Hi Kurt,
Sorry for the delay-- Have you tried the recent contrast-agnostic cord segmentation and the TotalSpineSeg vertebral labeling methods? I expect them to be more robust. Code would look like this (make sure you run SCT v7.1 or higher)
# This code has been tested with SCT v7.1
# Download example data
sct_download_data -d sct_example_data -o sct_example_data
cd sct_example_data/t1
# Crop to simulate a brain-only scan with the cord visible down to C2-C3 disc
sct_crop_image -i t1.nii.gz -zmin 125
# Segment the spinal cord, specifying that a brain is present in the image
sct_deepseg spinalcord -i t1_crop.nii.gz -qc qc
# Label vertebrae
sct_deepseg totalspineseg -i t1_crop.nii.gz -step1-only 1
sct_qc -i t1_crop.nii.gz -s t1_crop_step1_levels.nii.gz -p sct_label_utils -qc qc
# Compute average CSA between C1-C2 level
sct_process_segmentation -i t1_crop_seg.nii.gz -discfile t1_crop_step1_levels.nii.gz -vert 1:2
This has worked nicely on both cases above! Tested locally using SCT v7.1. I’m now building an apptainer with spinal cord and totalspineseg tasks (to run on our cluster) and will let you know what this looks like on a larger scale. Thank you!
Dear @schillkg,
I am very glad to hear that you are trying out the new-to-v7.1 Apptainer installation instructions. As this is a new installation method for SCT, if you run into any issues, please feel to open a new forum post and tag me, and I will be happy to help. 
Kindly,
Joshua
1 Like