dear jcohenadad , I just put the my patient’s dti imaging to the dmri document , and the other documents stay the same that I downloaded from the course ,it it will affect the accuracy of my patient’s dti?
it means only the imaging in dmri document is belong to my patient, the imanging in other document don’t belong to my patient. if it affects the accuracy of my patient’s dti in dmri document.
thank you .
i’m really sorry but i still don’t understand your question.
I want to explain , that I just put my patient’s dti imaging to the dmri document, and the other document is other patient’s(I don’t know who) imaging.( fmri, t1 ,t2 and so on). if I could get the right dti result of my patient?
my English is poor, please forgive me .
what do you mean by “I just put my patient’s dti imaging to the dmri document”. Did you mean folder? (instead of document)
if that’s what you meant, then you should not do that as the other images (t1, t2, etc.) are not from the same patient, and these data are used to process your dti data (e.g., warping field, etc.).
dear jcohenadad, thank you for your reply.
I think I have understood the sct.
1.I have the question. I have only one t1 imaging of c1-c7. however ,I have 3 t2 imaging (c1-c3,c3-c5,c5-c7),and 3 dti imaging (c1-c3,c3-c5,c5-c7). if I could put t1(c1-c7) in t1 folder, and t2(c1-c3) in t2 folder, dti(c1-c3) folder to calculate the dti (c1-c3) data ?
2. I think I have no the imaging of t2s ,what should I do? these are all imaging I have.
thank you .
hi,
you don’t absolutely need the T1 or T2 scan to compute DTI data. You could also register the PAM50 to your DTI scans without it, if you know the vertebral level of your DTI scans.
Now, if you want to use an anatomical image, you can either use the T1 or the T2 image, depending which one has the best spatial resolution (especially axial resolution). All these aspects are explained in the SCT course.
Regarding the 2nd question: as mentioned above, you do not need to have the T2s data in order to quantify the DTI data.
Best,
Julien
thank you ,
sct_register_multimodal -i $SCT_DIR/data/PAM50/template/PAM50_t1.nii.gz -iseg $SCT_DIR/data/PAM50/ template/PAM50_cord.nii.gz -d dmri_crop_moco_dwi_mean.nii.gz -dseg dmri_crop_moco_dwi_mean_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,slicewise=1,iter=3 -initwarp …/ t2s/warp_template2t2s.nii.gz -initwarpinv …/t2s/warp_t2s2template.nii.gz -qc ~/qc_singleSubj
This script need the t1 and t2, could you tell me how to avoid this?
First of all, this syntax does not require a T1w or T2w image. It is using the template (PAM50_t1.nii.gz). Secondly, you can avoid relying on an initial warping field (here a T2*w-based field) by running sct_register_to_template directly on your DTI image, as I mentioned above. Examples are provided in the course material, or on the sct-pipeline repos.
Dear jcohenadad
When I put the dti imaging into the t1 folder and t2 folder to make a segementation. However when it came to this step sct_propseg -i t2.nii.gz -c t2 -qc ~/qc_singleSubj(I replace the t2 with dti. and I change the name of dti.nii to the t2. nii) Then it shows ERROR: your input image needs to be 3D in order to be segmented. What is the problem?
The problem is that you input a 4d image. You need to average your dmri series across time before inputting it into propseg, as exemplified in the SCT course.
At this point it will be easier if I just write the batch script for your analyses instead.
Just let me know what metrics you want, send me an example dataset and I’ll provide the script for you
TANG_WING_KUN_WIP_DTI_thk7_breath_1_5_1.zip (738.2 KB)
Dear jcohenadad , thank you very much.
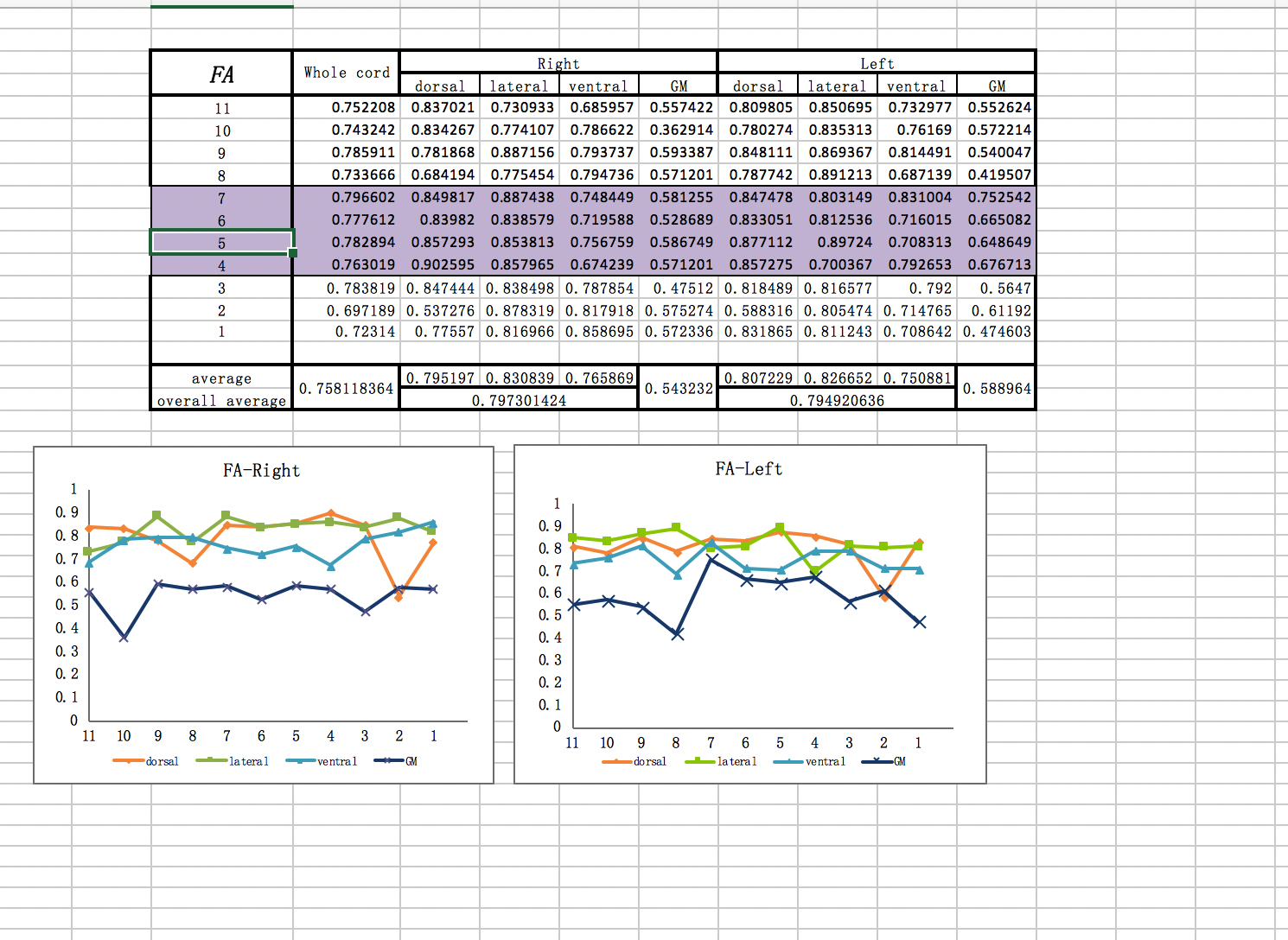
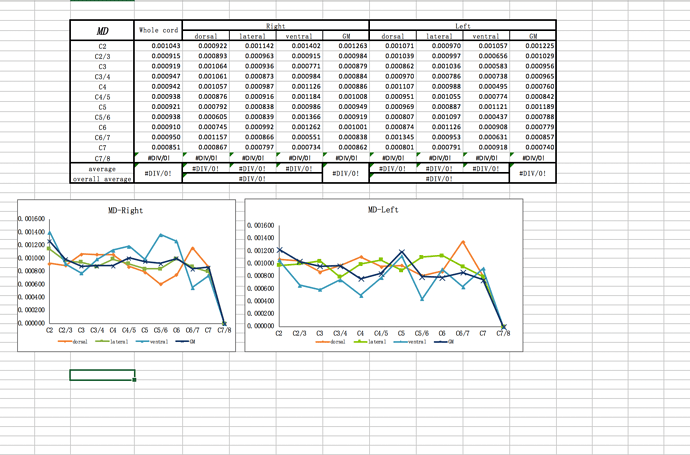
I want to get the patient’s gray matter‘s dti data , MD ,FA,RD value through this imaging I uploaded.
Thank you a lot!
Dear jcohenadad
I am sorry to bother you so long time , but I met too much problems before , what the data I want to get just like the picture I uploaded . these are the data of C1-C7 , the data I give you is C1-C3. but I think if I could deal with the DTI of C1-C3, it is enough for me.
Thank you.
Dear jcohenadad, maybe I could deal with that with your guidance.
hi,
sorry i am extremely busy these days. i was planning to work on your project probably sometimes next week. So, if I understand correctly: are you saying that you don’t need my help anymore?
dear jcohenadad, I think you are busy , but if you have time , I wish you could help me .
because it is too hard for me .
waiting for your time. thank you very much.
I’ve looked at your diffusion MRI data. In order to register it with the PAM50 template, I would need to know what level (disc or mid-vertebral body) the volume is centered at.
Dear Jcohenadad. Thank you so much for your attention. This data is C5-C7.
Thank you.
Here is the script to process your data:
# Create variable name for convenience
file_dwi="TANG_WING_KUN_WIP_DTI_thk7_breath_1_5_1"
file_bvec=${file_dwi}.bvec
file_bval=${file_dwi}.bval
# Separate b=0 and DW images
sct_dmri_separate_b0_and_dwi -i ${file_dwi}.nii.gz -bvec ${file_bvec}
# Segment cord (1st pass -- just to get a rough centerline)
sct_propseg -i ${file_dwi}_dwi_mean.nii.gz -c dwi
# Create mask to help motion correction and for faster processing
sct_create_mask -i ${file_dwi}_dwi_mean.nii.gz -p centerline,${file_dwi}_dwi_mean_seg.nii.gz -size 30mm
# Crop data for faster processing
sct_crop_image -i ${file_dwi}.nii.gz -m mask_${file_dwi}_dwi_mean.nii.gz -o ${file_dwi}_crop.nii.gz
# Motion correction
sct_dmri_moco -i ${file_dwi}_crop.nii.gz -bvec ${file_dwi}.bvec -x spline
file_dwi=${file_dwi}_crop_moco
file_dwi_mean=${file_dwi}_dwi_mean
# Segment spinal cord (only if it does not exist)
sct_deepseg_sc -i ${file_dwi_mean}.nii.gz -c dwi -qc qc
file_dwi_seg=${file_dwi_mean}_seg
# Create labels, assuming that the mid-FOV is centered at C6 vertebral level
sct_label_utils -i ${file_dwi_seg}.nii.gz -create-seg -1,6 -o label_C6.nii.gz
# Register DWI data to the PAM50 template
sct_register_to_template -i ${file_dwi_mean}.nii.gz -s ${file_dwi_seg}.nii.gz -l label_C6.nii.gz -ref subject -c t1 -param step=1,type=seg,algo=centermassrot:step=2,type=im,algo=syn,metric=CC,slicewise=0,smooth=0,iter=3 -qc qc
# Rename warping field for clarity
mv warp_template2anat.nii.gz warp_template2dwi.nii.gz
mv warp_anat2template.nii.gz warp_dwi2template.nii.gz
# Warp template
sct_warp_template -d ${file_dwi_mean}.nii.gz -w warp_template2dwi.nii.gz -qc qc
# Compute DTI
sct_dmri_compute_dti -i ${file_dwi}.nii.gz -bvec ${file_bvec} -bval ${file_bval} -method standard
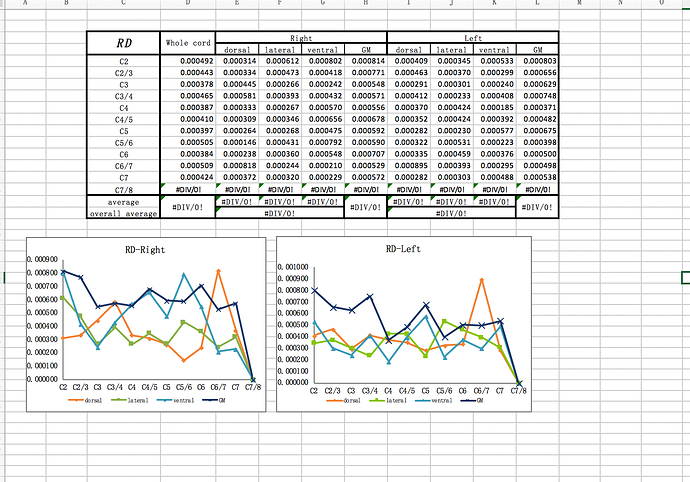
# Compute FA, MD and RD in WM and GM between C5 and C7 vertebral levels
sct_extract_metric -i dti_FA.nii.gz -f label/atlas -l 51,52 -vert 5:7 -o DWI_FA.csv -append 1
sct_extract_metric -i dti_MD.nii.gz -f label/atlas -l 51,52 -vert 5:7 -o DWI_MD.csv -append 1
sct_extract_metric -i dti_RD.nii.gz -f label/atlas -l 51,52 -vert 5:7 -o DWI_RD.csv -append 1
And here are the results (QC report and csv files): results.zip (382.8 KB)
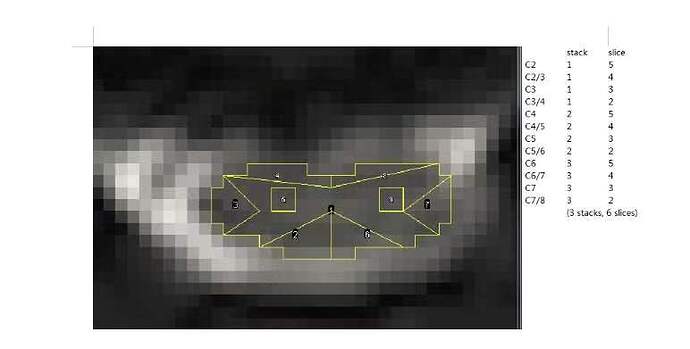
dear jcohenadad, I have a question . if the sct could get the ROI just like this? it is our team’s aim.
thank you . if you can, could you help me. I am sorry for bothering you a long time.
hi,
if you look at the usage of sct_extract_metric, you will notice that the labels can be called (via the flag -l). Here is a list of combined labels that you might find relevant:
50, spinal cord, 0:35
51, white matter, 0:29
52, gray matter, 30:35
53, dorsal columns, 0:3
54, lateral funiculi, 4:13
55, ventral funiculi, 14:29
and if you want, let’s say, to separate the right and left funiculi, you could edit the file info_label.txt (as instructed here) and add the following entries:
56, left funiculi, 4,6,8,10,12
57, right funiculi, 5,7,9,11,13
and then run sct_extract_metric with these new labels.
best,
Julien