Dear SCT Team:
When I was doing gray matter segmentation, the results I got were not ideal.
The command I used is as follows:"sct_deepseg_gm -i t2_te_13.7_reg.nii.gz -qc ~/qc_singleSubj
And this is the data:
t2s_te_13.7_reg.nii.zip (9.5 MB)
Dear @ryu,
Thank you for your question, and for sharing sample data to help us debug. ![]()
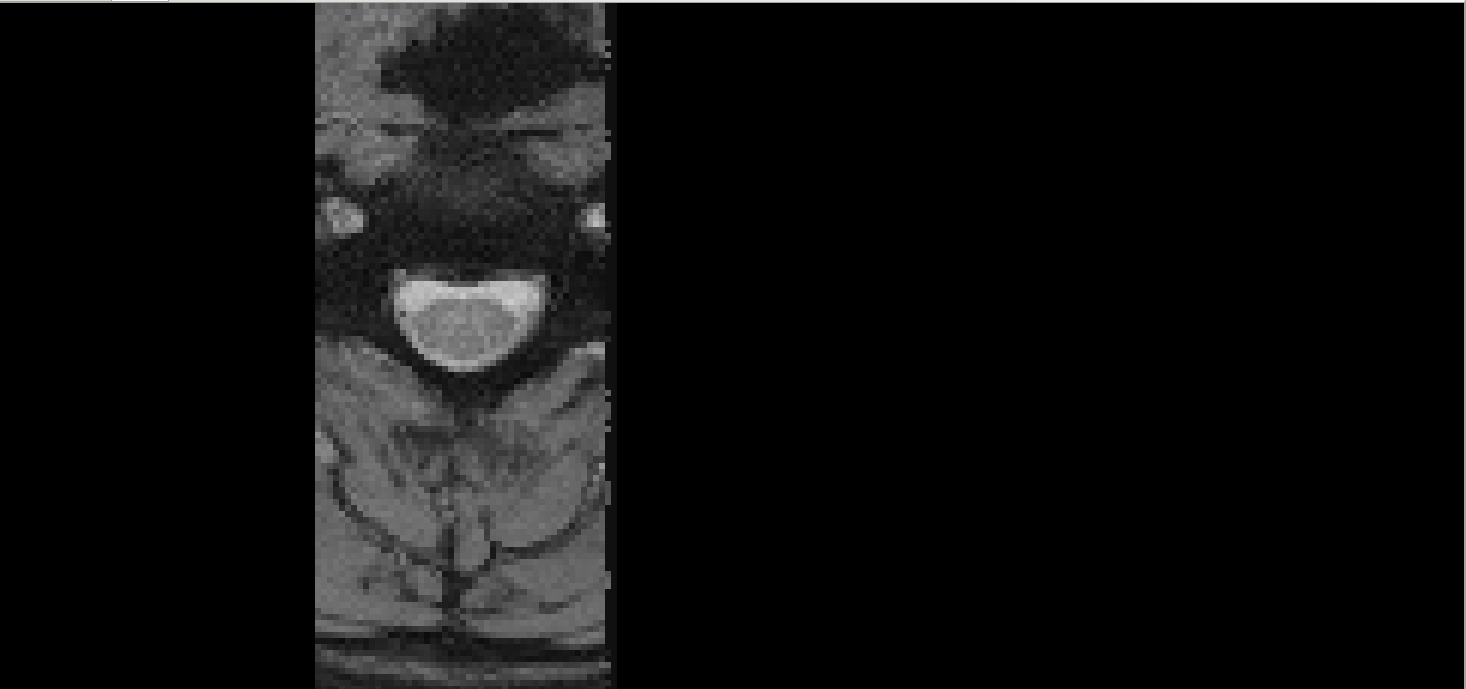
The issue is most likely due to data quality. There appears to be insufficient contrast between the white matter and gray matter. For example, on axial slice 150:
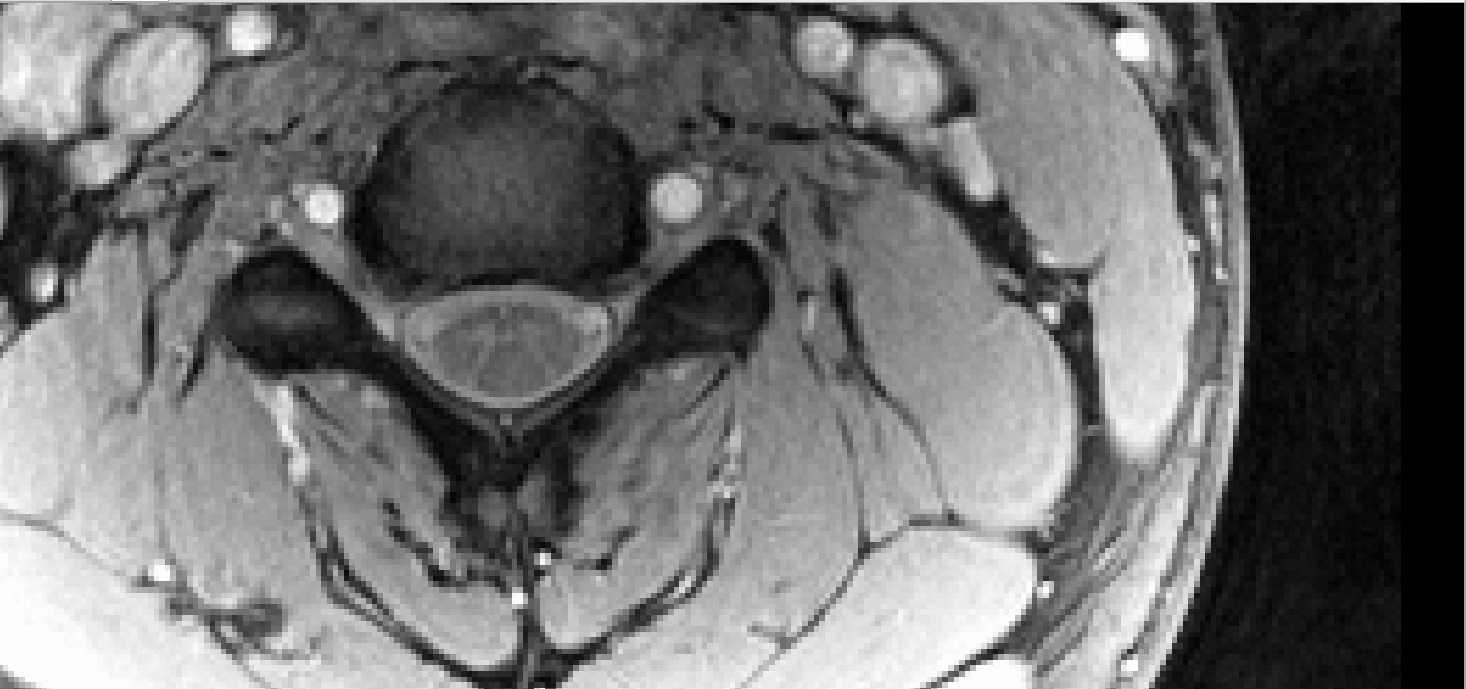
Compare to the sample data used by SCT to test gray matter segmentation:
I have tried using cropping to improve the segmentation quality, as described in this previous forum post:
sct_deepseg_sc -i t2s_te_13.7_reg.nii.gz -c t2s
sct_crop_image -i t2s_te_13.7_reg.nii.gz -m t2s_te_13.7_reg_seg.nii.gz -dilate 10
sct_deepseg_gm -i t2s_te_13.7_reg_crop.nii.gz
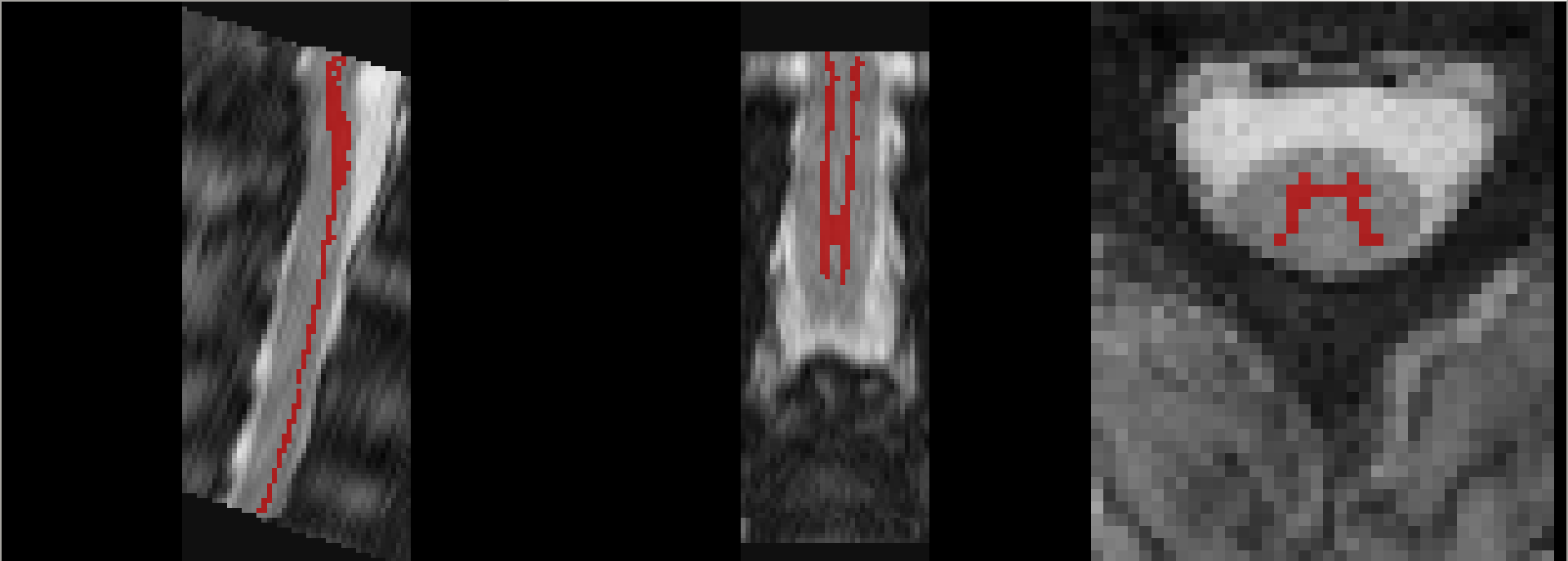
The results are a bit better, but still not 100% ideal:
This could be a good starting point for further manual corrections, though. ![]()
Kind regards,
Joshua
Please note that sct_deepseg_gm was also designed for T2* data with thick axial slices. For example, our test data has 32 axial slices, with each slice having size 320x320 (i.e. fewer high-resolution slices), while your data has 320 axial slices, with each slice having size 50x320 (i.e. many low-resolution slices).
Due to the differences in axis order (320x320x32 vs. 50x320x320), I’m wondering if there are differences in how your data was acquired compared to our test data. (e.g. axially vs. sagittally) ![]()
((Also, note that because there are 320 axial slices, this is what is causing the large memory usage during inference: Tensorflow-based ONNX models using a lot of memory on large image · Issue #4612 · spinalcordtoolbox/spinalcordtoolbox · GitHub))
Thank you very much!