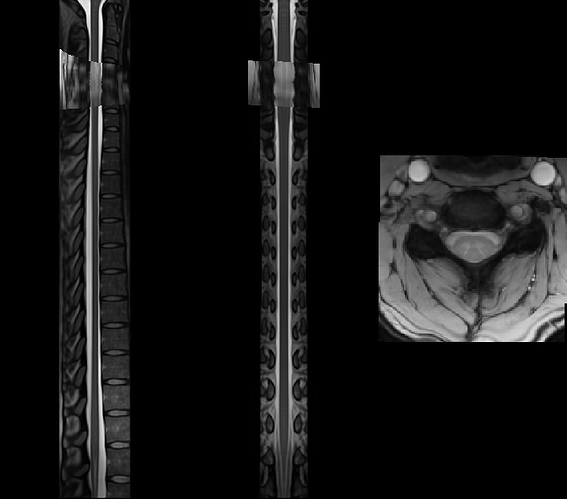
Hi,
I am pretty sure there is no problem with the viewer, as I tried to visualize it with multiple viewers.
Here is my script:
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Wednesday 24 may 2023
@author: evefra
"""
# import libraries
import os
import shutil
import subprocess
import fnmatch
import nibabel as nib
import numpy as np
# work in conda env, where SCT is installed. For me: conda activate venv_sct
######################################################################################
# INDICATE WHAT TO DO#
######################################################################################
# ANATOMICAL T1
first_segment_t1 = 0
smooth = 0
second_segment_t1 = 0 # Correct
label_t1 = 0
register_t1_temp = 1 # NOT CORRECT
# LOOP OVER SUB NUMBERS
# ------------------------------------------------------------------------------
sub_list = [20]
for sub in sub_list:
# data path pre subject unspecific
data_pre = "/project/3023009.09/evefra"
subdirectory = f"sub-{sub:03d}"
#################################################################################
# T1 prep #
#################################################################################
# FIRST SEGMENTAION OF T1 (including manual C2 and T1 labelling)
# ------------------------------------------------------------------------
if first_segment_t1 == 1:
# define input
input_directory = os.path.join(
data_pre, subdirectory, "anat")
wildcard_pattern = 's*.nii'
input_file = fnmatch.filter(os.listdir(
input_directory), wildcard_pattern)
input_path = os.path.join(input_directory, input_file[0])
# define output
output_directory = os.path.join(
data_pre, subdirectory, "spinalcord", "segment_anat")
if not os.path.exists(output_directory):
os.makedirs(output_directory)
output_file = input_file[0].replace("nii", "segmentation.nii")
output_path = os.path.join(output_directory, output_file)
# define QC directory
qc_directory = os.path.join(output_directory, 'qc')
# run first segmentation
# label centerline C2 and C7
command = f"sct_deepseg_sc -i {input_path} -c t1 -o {output_path} -qc {qc_directory} -centerline viewer"
process = subprocess.Popen(command, shell=True)
process.wait()
print('first segment t1 completed for ', subdirectory)
# SMOOTH ALONG SC TO IMPROVE SEGMENTATION
# -------------------------------------------------------------------------------
if smooth == 1:
# define input
input_directory = os.path.join(
data_pre, subdirectory, "anat")
wildcard_pattern = 's*.nii'
input_file = fnmatch.filter(os.listdir(
input_directory), wildcard_pattern)
input_path = os.path.join(input_directory, input_file[0])
# select segmentation image
input_directory_seg = os.path.join(
data_pre, subdirectory, 'spinalcord', "segment_anat")
wildcard_pattern = '*.segmentation.nii'
input_file_seg = fnmatch.filter(os.listdir(
input_directory_seg), wildcard_pattern)
input_path_seg = os.path.join(input_directory_seg, input_file_seg[0])
# define output
output_directory = os.path.join(
data_pre, subdirectory, "spinalcord", "segment_anat")
output_file_seg = input_file_seg[0].replace(
"segmentation.nii", "smooth.nii")
output_path_seg = os.path.join(output_directory, output_file_seg)
# RUN SMOOOTHING
command = f"sct_smooth_spinalcord -i {input_path} -o {output_path_seg} -s {input_path_seg} -smooth 0,0,5"
process = subprocess.Popen(command, shell=True)
process.wait()
print('smooth t1 completed for ', subdirectory)
# DO SECOND SEGMENTATION
# -----------------------------------------------------------------------------
if second_segment_t1 == 1:
# define input
input_directory = os.path.join(
data_pre, subdirectory, "spinalcord", "segment_anat")
wildcard_pattern = '*smooth.nii'
input_file = fnmatch.filter(os.listdir(
input_directory), wildcard_pattern)
input_path = os.path.join(input_directory, input_file[0])
# define output
output_directory = os.path.join(
data_pre, subdirectory, "spinalcord", "segment_anat_second")
if not os.path.exists(output_directory):
os.makedirs(output_directory)
output_file = input_file[0].replace(
"smooth.nii", "second_segmentation.nii")
output_path = os.path.join(output_directory, output_file)
# define qcdirectory
qc_directory = os.path.join(output_directory, 'qc')
# select segmentation image
input_directory_seg = os.path.join(
data_pre, subdirectory, 'spinalcord', "segment_anat")
wildcard_pattern = '*.segmentation.nii'
input_file_seg = fnmatch.filter(os.listdir(
input_directory_seg), wildcard_pattern)
input_path_seg = os.path.join(input_directory_seg, input_file_seg[0])
# run second segmentation
command = f"sct_deepseg_sc -i {input_path} -c t1 -o {output_path} -qc {qc_directory} -centerline file -file_centerline {input_path_seg}"
process = subprocess.Popen(command, shell=True)
process.wait()
print('second segment t1 completed for ', subdirectory)
# CHECK SEGMENTATION AND CORRECT
# --------------------------------------------------------------------------------
# in FSLeyes
# LABEL T1
# -------------------------------------------------------------------------------
if label_t1 == 1:
# define input anatomical image
input_directory = os.path.join(
data_pre, subdirectory, "anat")
wildcard_pattern = 's3*-01.nii'
input_file = fnmatch.filter(os.listdir(
input_directory), wildcard_pattern)
input_path = os.path.join(input_directory, input_file[0])
# define input segmentation
input_directory_seg = os.path.join(
data_pre, subdirectory, 'spinalcord', "segment_anat_second")
wildcard_pattern = '*segmentation.nii'
input_file_seg = fnmatch.filter(os.listdir(
input_directory_seg), wildcard_pattern)
input_path_seg = os.path.join(input_directory_seg, input_file_seg[0])
# define output paths and name
output_directory = os.path.join(
data_pre, subdirectory, "spinalcord", "label_anat")
if not os.path.exists(output_directory):
os.makedirs(output_directory)
output_file_manual = input_file[0].replace(".nii", "manlabel.nii")
output_path_manual = os.path.join(output_directory, output_file_manual)
output_file = input_file[0].replace(".nii", "label.nii")
output_path = os.path.join(output_directory, output_file)
# define qc directory
qc_directory_man = os.path.join(output_directory, 'qc_manual')
qc_directory = os.path.join(output_directory, 'qc')
# manual label C2 and C7
command = f"sct_label_utils -i {input_path} -qc {qc_directory_man} -create-viewer 3,7 -o {output_path_manual} -msg 'Please manually label the inter-vertebral disc '"
process = subprocess.Popen(command, shell=True)
process.wait()
# define input man label
# wildcard_pattern = '*manlabel.nii'
# input_file_man = fnmatch.filter(os.listdir(
# output_directory), wildcard_pattern)
# input_path_man = os.path.join(output_directory, input_file_man[0])
# correcting s or qform if needed
# sct_image -set-sform-to-qform -i {
# run labelling
# command = f"sct_label_vertebrae -i {input_path} -c t1 -s {input_path_seg} -qc {qc_directory} -o {output_path} -initlabel {input_path_man}"
# process = subprocess.Popen(command, shell=True)
# process.wait()
print('label t1 completed for ', subdirectory)
# REGISTER FROM T1 to TEMPLATE
# -----------------------------------------------------------------------------
if register_t1_temp == 1:
# define input image (T1)
input_directory = os.path.join(
data_pre, subdirectory, "anat")
wildcard_pattern = 's3*01.nii'
input_file = fnmatch.filter(os.listdir(
input_directory), wildcard_pattern)
input_path = os.path.join(input_directory, input_file[0])
# define input segmentation
input_directory_seg = os.path.join(
data_pre, subdirectory, 'spinalcord', "segment_anat_second")
wildcard_pattern = '*segmentation.nii'
input_file_seg = fnmatch.filter(os.listdir(
input_directory_seg), wildcard_pattern)
input_path_seg = os.path.join(input_directory_seg, input_file_seg[0])
# define labels input image
input_directory_lab = os.path.join(
data_pre, subdirectory, "spinalcord", "label_anat")
wildcard_pattern = 's*manlabel.nii' # name if not manual labelled?
input_file_lab = fnmatch.filter(os.listdir(
input_directory_lab), wildcard_pattern)
input_path_lab = os.path.join(input_directory_lab, input_file_lab[0])
# define output directory
output_directory = os.path.join(
data_pre, subdirectory, "spinalcord", "register_t1_temp")
# define qc
qc_directory = os.path.join(output_directory, 'qc_t1_temp')
# run registering T1 --> template
command = f"sct_register_to_template -i {input_path} -s {input_path_seg} -l {input_path_lab} -c t1 -qc {qc_directory} -ofolder {output_directory}"
process = subprocess.Popen(command, shell=True)-
process.wait()
print('register calc t1 - temp completed for ', subdirectory)
I will send the data private to you.
Thank you in advance.
Best,
Evelyne
![[animate output image]](https://im3.ezgif.com/tmp/ezgif-3-56d20faabc.gif)
![[animate output image]](https://im3.ezgif.com/tmp/ezgif-3-156dc0eafc.gif)