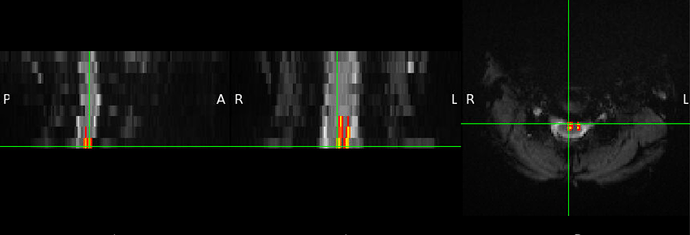
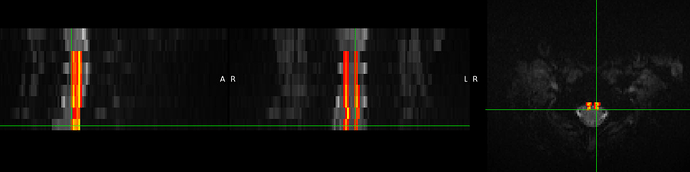
Thankyou @joshuacwnewton
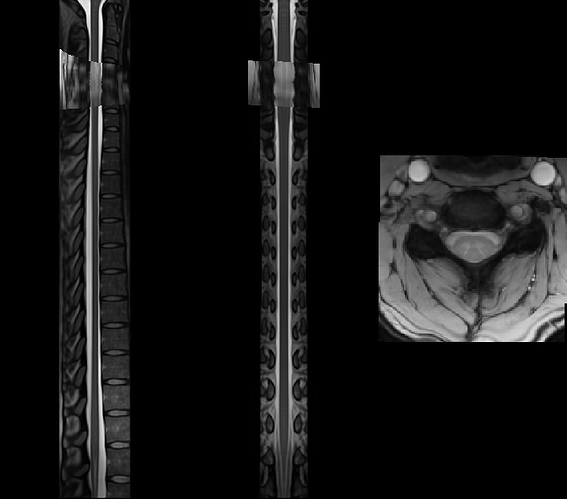
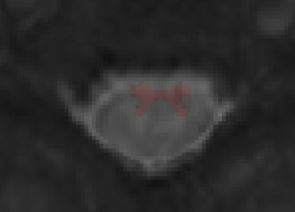
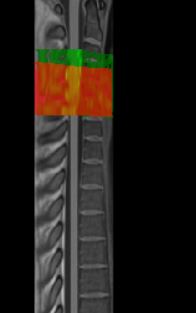
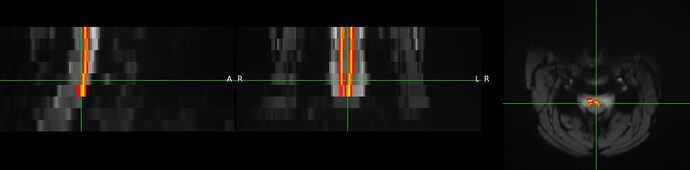
Did you already figure out why a blank screen occurred? I get another blank screen when improving my registration using sct_multimodal_transfo and gm segmentations. Without the addition of gm, the warping field is working. Some data (t2, initial warp, my result): SURFdrive
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Wednesday 24 may 2023
@author: evefra
"""
# import libraries
import os
import shutil
import subprocess
import fnmatch
import nibabel as nib
import numpy as np
# work in conda env, where SCT is installed. For me: conda activate venv_sct
######################################################################################
# INDICATE WHAT TO DO#
######################################################################################
# COREGISTERING
register_t2_t1 = 0
register_fmri_t2 = 0
t2_to_t1_mult_t1_to_temp = 0
correct_qform_sc = 0
gm_segment = 0
gm_segment_add = 0
epi_to_t2_mult_t2_to_temp = 0
final_warp_apply = 0
# LOOP OVER SUB NUMBERS
# ------------------------------------------------------------------------------
sub_list = [10]
# [1, 2, 4, 5, 6, 8, 9, 10, 11, 13, 14, 16, 17, 18, 24]
for sub in sub_list:
# CORRECT QFORM ANATOMICAL
# -----------------------------------------------------------------------------
if correct_qform_sc == 1:
# Define the input
input_directory_d = os.path.join(
data_pre, subdirectory, "anat_sc")
wildcard_pattern_d = 's3*01.nii'
input_file_d = fnmatch.filter(os.listdir(
input_directory_d), wildcard_pattern_d)
input_path_des = os.path.join(input_directory_d, input_file_d[0])
# define output
output_directory = os.path.join(
data_pre, subdirectory, "anat_sc")
output_file = input_file_d[0].replace("nii", "qform.nii")
output_path = os.path.join(output_directory, output_file)
# run command
command = f"sct_image -i {input_path_des} -set-sform-to-qform -o {output_path}"
process = subprocess.Popen(command, shell=True)
process.wait()
print('qform fix anat sc completed for ', subdirectory)
# CALC SEGMENTATION GM
# ----------------------------------------------------------------------------
if gm_segment == 1:
# define input
input_directory_d = os.path.join(
data_pre, subdirectory, "anat_sc")
wildcard_pattern_d = 's3*01.qform.nii'
input_file_d = fnmatch.filter(os.listdir(
input_directory_d), wildcard_pattern_d)
input_path_des = os.path.join(input_directory_d, input_file_d[0])
# define output
output_directory = os.path.join(
data_pre, subdirectory, "spinalcord", "gm_segment_t2_anat")
output_file_seg = 'gm_t2.nii'
output_path = os.path.join(output_directory, output_file_seg)
# Check if the removed volumes directory exists, create it if necessary
if not os.path.exists(output_directory):
os.makedirs(output_directory)
# define qc
qc_directory = os.path.join(output_directory, 'qc')
# Run gm segment
command = f"sct_deepseg_gm -i {input_path_des} -qc {qc_directory} -o {output_path}"
process = subprocess.Popen(command, shell=True)
process.wait()
print('segment gm completed for ', subdirectory)
# ADD SEGMENTATION GM
# ----------------------------------------------------------------------------
if gm_segment_add == 1:
# define input template (always the same)
input_directory = '/home/affneu/evefra/sct_5.8/data/PAM50/template/PAM50_t1.nii.gz'
# define input gm segmentation (always the same)
input_directory_iseg = '/home/affneu/evefra/sct_5.8/data/PAM50/template/PAM50_gm.nii.gz'
# define destination (T2)
input_directory_d = os.path.join(
data_pre, subdirectory, "anat_sc")
wildcard_pattern_d = 's3*01.nii'
input_file_d = fnmatch.filter(os.listdir(
input_directory_d), wildcard_pattern_d)
input_path_des = os.path.join(input_directory_d, input_file_d[0])
# define destination segmentation (T2 GM)
input_directory_des_seg = os.path.join(
data_pre, subdirectory, "spinalcord", "gm_segment_t2_anat")
wildcard_pattern_des_seg = 'gm*.nii'
input_file_des_seg = fnmatch.filter(os.listdir(
input_directory_des_seg), wildcard_pattern_des_seg)
input_path_des_seg = os.path.join(
input_directory_des_seg, input_file_des_seg[0])
# define init warp
input_directory_warp = os.path.join(
data_pre, subdirectory, "spinalcord", "concat_t2_t1_temp")
wildcard_pattern_warp = 'warp_t22temp.nii.gz'
input_file_warp = fnmatch.filter(os.listdir(
input_directory_warp), wildcard_pattern_warp)
input_path_des_warp = os.path.join(
input_directory_warp, input_file_warp[0])
# define output warp
output_directory = os.path.join(
data_pre, subdirectory, "spinalcord", "concat_t2_t1_temp")
if not os.path.exists(output_directory):
os.makedirs(output_directory)
output_file = 'gm_warp_t22temp.nii.gz'
output_path_warp = os.path.join(output_directory, output_file)
print(input_directory)
print(input_directory_iseg)
print(input_path_des)
print(input_path_des_seg)
print(input_path_des_warp)
print(output_path_warp)
# run warping calculation
command = f"sct_register_multimodal -i {input_directory} -iseg {input_directory_iseg} -d {input_path_des} -dseg {input_path_des_seg} -initwarp {input_path_des_warp} -owarp {output_path_warp}"
process = subprocess.Popen(command, shell=True)
process.wait()
print('add gm to t2-temp warp completed for ', subdirectory)
![]()
![[animate output image]](https://im3.ezgif.com/tmp/ezgif-3-56d20faabc.gif)
![[animate output image]](https://im3.ezgif.com/tmp/ezgif-3-156dc0eafc.gif)